What is systematic review?
Systematic reviews collect and analyze documents to find answers to important questions. The #sysrev community is revolutionizing review by upgrading traditional techniques with modern web applications and artificial intelligence.

Systematic reviews collect and analyze documents to find answers to important questions. sysrev.com is revolutionizing review with a modern web application and artificial intelligence.
Sysrev.com is a free tool that merges open access principles into systematic literature review to create a new kind of fully reproducible literature review. Sysrev is already in use in hundreds of public projects - 15 of which match the query 'cancer' alone.
If you want to get started now, just visit the landing page and sign up - sysrev is free for public projects. You can also use our short tutorial to help you get started at blog.sysrev.com/getting-started.
In this post, we explain how systematic reviews work and how to perform them with the latest computational tools.
What is a systematic review?
Sysrevs can be broken into 4 steps:
- Set a clear Objective
- Search for relevant literature
- Screen literature and Extract data
- Report results
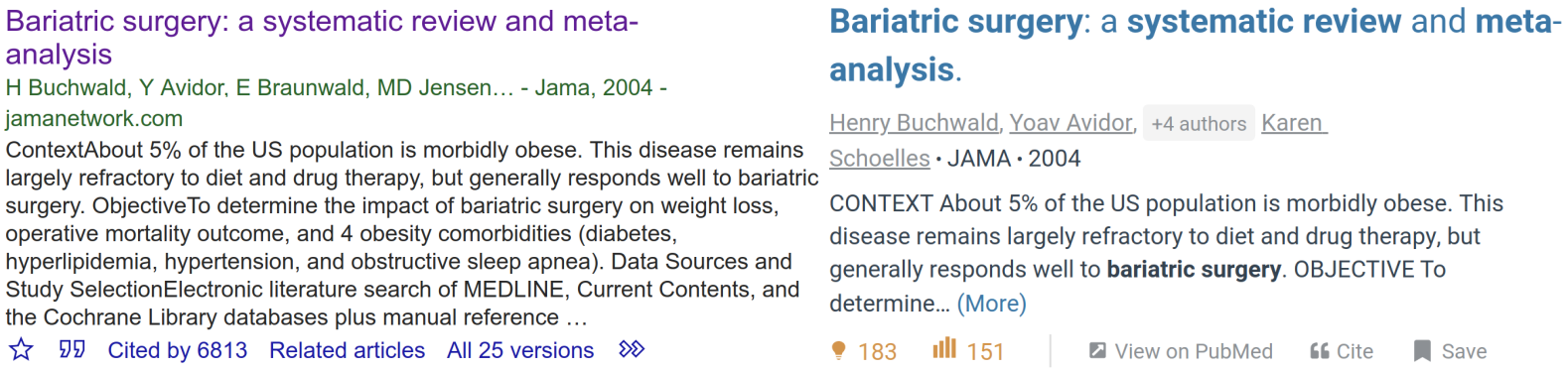
Objective: In a 2004 systematic review Bariatric Surgery - A Systematic Review and Meta-analysis Buchwald sought to "determine the impact of bariatric surgery on weight loss, operative mortality outcome, and 4 obesity comorbidities..."[1]. These 3 objectives clearly state the purpose of the review. If properly answered they may change the way bariatric surgery is performed.
In fact, the outcomes of these studies have significant impacts, the above publication has been referenced more than 6000 times making it one of the most authoritative sources on bariatric surgery.
Objectives are still very much a human endeavor. It helps to begin any project with a clear idea of what you want to achieve. Clear goals help to inform every other step in a review, and let you define whether or not you are advancing toward an end.
Some goals could eventually be automated, particularly goals that can be generalized. Questions like "What treatments are most effective against disease x?" and "What genetic variants cause adverse reactions to treatment z?" can be posed and tackled by natural language processing methods. Still these questions rely on some systematic understanding of adverse reactions, diseases, and treatments.
Naming and defining of the categories, properties and relations through formal Ontologies comes in useful here, but current review tools have only just begun this practice. Ontologies are a method of representing language in a way understandable by computers. The NCBI medical subject heading (MESH) database could be considered an ontology, and the OBO foundry contains many downloadable ontologies of diseases, drugs, genes, and more.
Search: To answer the objective, careful search queries must be formed to find documents relevant to the topic in an unbiased manner. Larger literature reviews will sometimes cite several medical literature databases. Buchwald's review uses MEDLINE, Current Contents, and the Cochrane Library to collect 2738 citations[1].
Review publications will sometimes provide a diagram detailing how many articles were reviewed. Unfortunately, it usually isn't possible to gain direct access to the collected citations in a traditional systematic review. However, computational tools can allow researchers to present greater transparency in their review workflow by releasing all the data generated during their review process. This includes what articles were collected.
By allowing reviewers to access all of the search data in a review it becomes easier for them to reference and expand upon previous work.
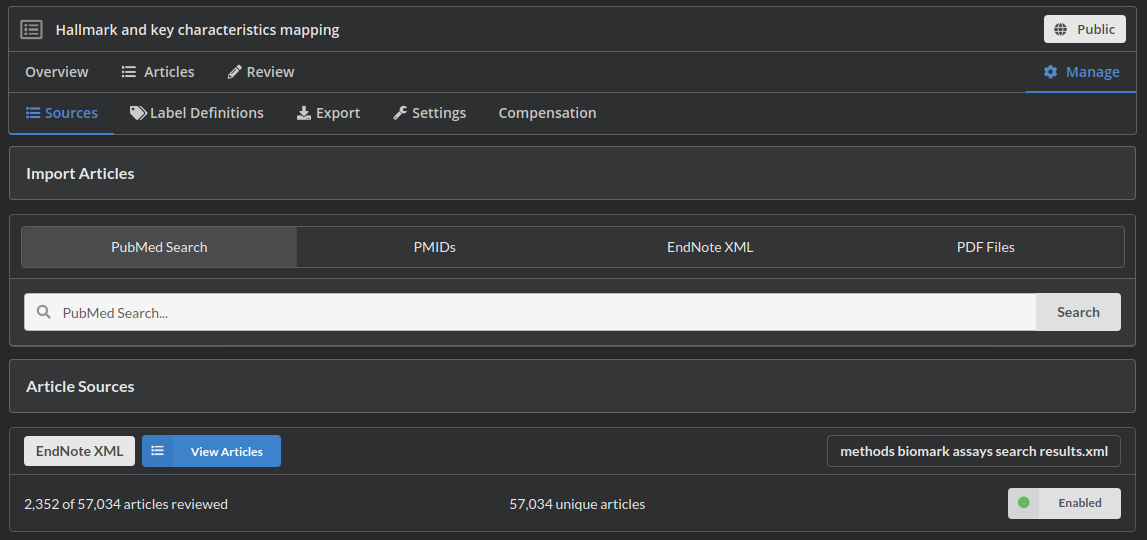
Screen/Extract: The process of actually reviewing documents involves both screening and extracting data. In the screening process articles are discarded when they fail to meet some well defined inclusion/exclusion criteria. During data extraction well defined questions are answered for the included articles.
Individual methods vary, but typically systematic reviews are performed collaboratively, with multiple people reviewing each document.
In her Coursera course on evidence based toxicology Dr. Lena Smirnova made the review method more educational by asking that all students review all documents (left below). The educational use of systematic review will receive more attention in future posts!
In the screening step for her liver toxicity review, Dr. Katya Tsaioun from the Evidence Based Toxicology Collaboration at Johns Hopkins required two reviewers per document (right below).

Tools like sysrev can help manage this screening and extraction process. At their most basic, they simply allot tasks to the relevant human reviewers.
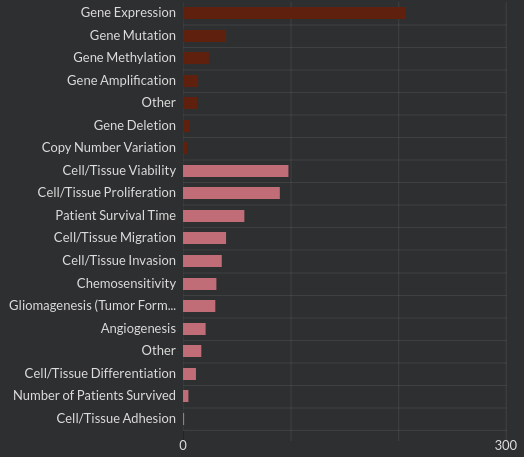
Data extraction is an extension of screening. In the extraction phase reviewers "enrich" the raw document data by identifying labels or annotating text. For example in a review on glioblastoma pathways researchers identified the type of glioma covered in each document (mesenchymal, proneural, classic or neural) along with the kind of data collected in the document (gene expression, mutation, methylation, amplication, copy number variation, other). This data extraction is extraordinarily laborious and frequently repeated in separate reviews. Computational tools will make it possible to share extracted data between systematic reviews.
Report: Once the review process is complete, data is collected and conclusions are drawn. In Buchwald's case, "Effective weight loss was achieved in morbidly obese patients after undergoing bariatric surgery. A substantial majority of patients with diabetes, hyperlipidemia, hypertension, and obstructive sleep apnea experienced complete resolution or improvement."[1] Sounds like bariatric surgery can have some major effects.
The report step of systematic reviews is usually delivered in a publication. The peer review process comes into play here and can help ensure good research. However, when reading the conclusions of a review, we must trust that the outcomes were correctly aggregated and without bias. Computational tools can help here as well. By making all of the data in a review open access reviewers can analyze the data themselves. Future posts will show programming APIs can allow for automated fact checking in systematic reviews.
Finality is a bigger problem with the report process. Every published review is never more correct than the day it is published (or arguably several months before it is published). This is because new publications occur over time and eventually the review becomes dated. Several authors have suggested 'living reviews' that update over time. Creation and use of the data in such living reviews will rely on computational tools.
We built sysrev.com to add transparency to the systematic review process and extend it to more fields.
What is a "sysrev"?
Sysrev is the world's first open access review platform. It hopes to extend the concept of systematic review to new data sources, automate the process of review, and democratize review with FAIR (findable, accessible, interoperable, reproducible) principles.
Projects on sysrev, affectionately called "sysrevs", can be made open access by marking them public. Public reviews can be easily found on google or on sysrev's own search engine. Search for "gene hunter sysrev" to find the gene hunter sysrev.
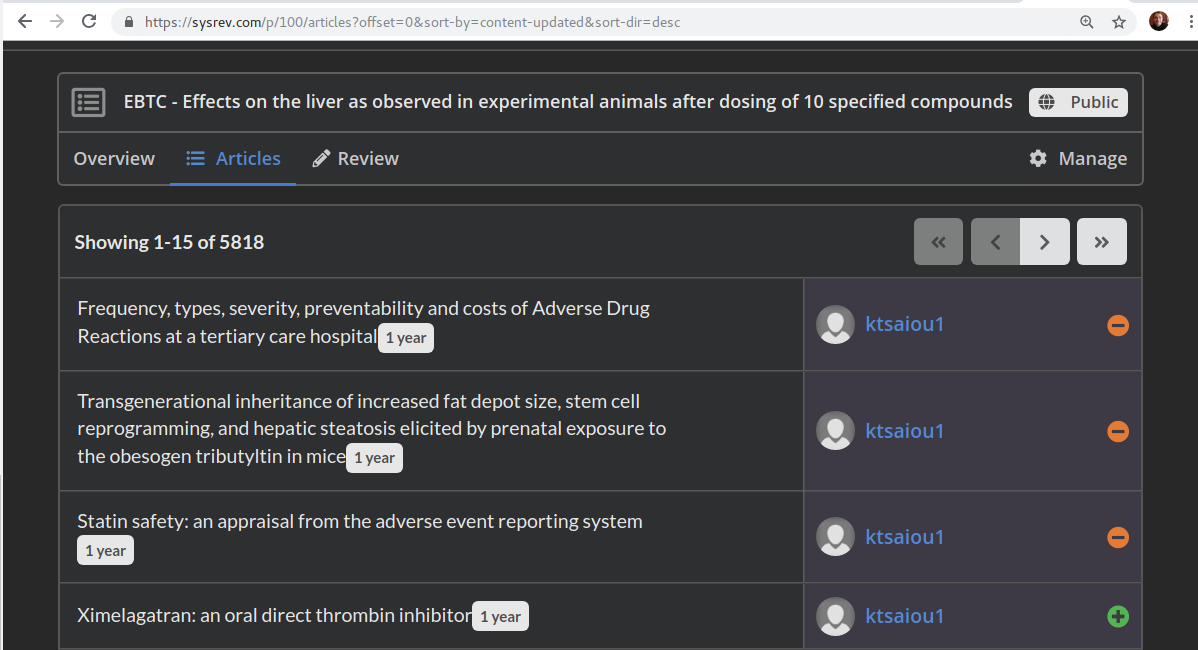
Users can also download data from public reviews and view detailed information on who reviewed which articles in a public review. For instance, you can visit the articles reviewed in the Hopkins sysrev on liver toxicity at sysrev.com/p/100. By clicking a user name, you can view their user page and see other projects on which they participated.
The systematic review process is a powerful way to extract actionable information from documents. Sysrev.com provides an open access platform to make the review process more transparent.
References
[1] Buchwald, Henry, et al. "Bariatric surgery: a systematic review and meta-analysis." Jama 292.14 (2004): 1724-1737.
